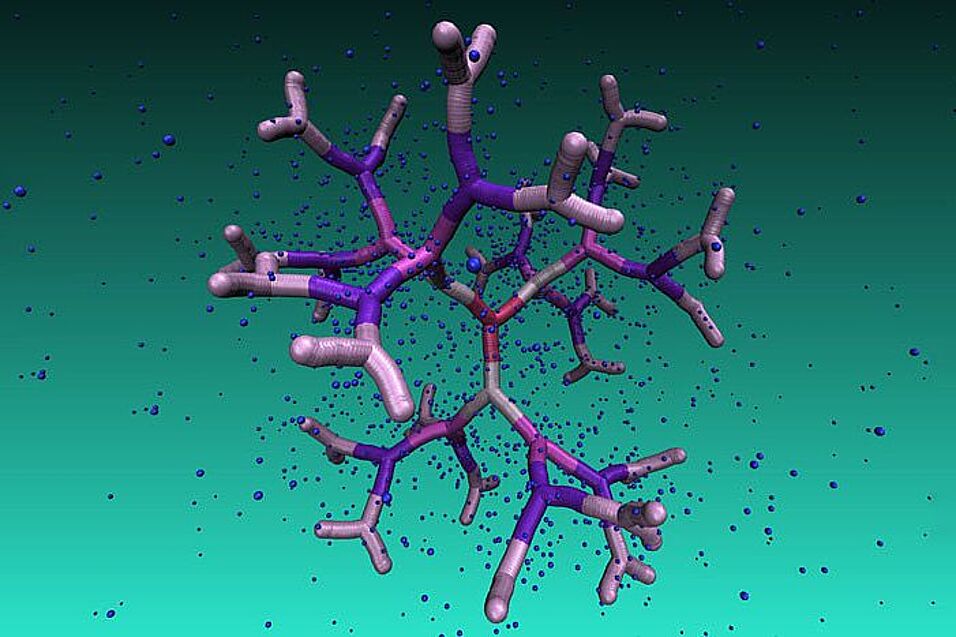
Nanokäfige sind hochinteressante molekulare Strukturen, sowohl aus der Sicht der Grundlagenforschung als auch in Hinblick auf mögliche Anwendungen. Die Hohlräume dieser nanometergroßen Objekte können als Träger kleinerer Moleküle genutzt werden, was in der Medizin für den Medikamenten- oder Gentransport in lebenden Organismen entscheidend ist. Diese Idee brachte ForscherInnen aus verschiedenen interdisziplinären Bereichen zusammen, die Dendrime – besondere chemische Verbindungen – als vielversprechende Kandidaten für die Herstellung solcher Nanoteilchen-Träger untersuchen. Die baumartige Architektur der Dendrimere und ihr schrittweises Wachstum mit sich wiederholenden, selbstähnlichen Einheiten erlauben die Ausformung von Hohlräumen mit kontrollierbarem Design. Jahrzehntelange Forschungen haben jedoch gezeigt, dass eine Vielzahl von verschiedenen Dendrimer-Arten mit zunehmenden Dendrimergenerationen eine Rückfaltung der äußeren Äste erfahren, was zu einer höheren Dichte der Bestandteile im Inneren des Moleküls führt. Die Wirkung des Rückfaltens wird durch Zugabe von Salz in die Lösung verstärkt, wodurch flexible Dendrimere stark schrumpfen und zu kompakten Objekten ohne Hohlräume in deren Innerem werden.
Das Team um Nataša Adžić und Christos Likos von der Universität Wien, Clemens Jochum und Gerhard Kahl (TU Wien), Emmanuel Stiakakis (Forschungszentrum Jülich, Deutschland) und Thomas Derrien und Dan Luo (Cornell University, USA) fand einen Weg, Dendrimere zu erzeugen, die so starr sind, dass eine Rückfaltung der äußeren Arme auch bei hohen Verzweigungsgenerationen verhindert wird. Somit bleiben regelmäßige Hohlräume in ihrem Inneren erhalten. Darüber hinaus zeichnen sich die neuartigen Makromoleküle durch eine bemerkenswerte Resistenz gegen Salzzusatz aus: Die WissenschafterInnen zeigten, dass die Morphologie und Konformationseigenschaften dieser Systeme auch bei Zugabe von Salz selbst in hoher Konzentration unbeeinflusst bleiben.
Die Nanokäfige, die sie im Labor und am Computer erzeugten, sind DNA-basierte Dendrimere, sogenannte Dendrimer-ähnliche DNAs (DL-DNA). Der Baustein, aus dem sie bestehen, ist eine Y-förmige doppelsträngige DNA-Einheit, eine dreiarmige Struktur aus doppelsträngiger DNA (ds-DNA). Diese wird durch Hybridisierung von drei einzelsträngigen DNA-Ketten (ss-DNA), die jeweils teilweise komplementäre Sequenzen zu den beiden anderen aufweisen, gebildet. Jeder Arm besteht aus 13 Basenpaaren und einem einzelsträngigen Klebeende mit vier Nukleinbasen, welches als Klebstoff fungiert. Während eine einzelne Y-DNA der ersten Dendrimer-Generation entspricht, ergibt das Anhängen weiterer Y-DNA-Elemente DL-DNA höherer Generationen. Das resultierende Dendrimer ist eine geladene makromolekulare Anordnung mit Hohlräumen und baumartiger Architektur. Aufgrund der Steifigkeit der dsDNA sind die Zweige der DL-DNA ziemlich starr, so dass das gesamte Molekül starr ist. Da DNA geladen ist, erhöht die elektrostatische Abstoßung zusätzlich die Steifigkeit des Moleküls.
DL-DNA-Moleküle wurden im Labor von den PartnerInnen in Jülich und Cornell mit bemerkenswerter Kontrolle und Sub-Nanometer-Präzision durch programmierbare Klebeende-Kohäsionen zusammengesetzt. Ihr schrittweises Wachstum ist in hohem Maß kontrollierbar, unidirektional und nicht umkehrbar. Diese Eigenschaft ist von großer Bedeutung, da gezeigt werden konnte, dass DNA-basierte Dendrimere eine vielversprechende Rolle bei der Entwicklung von nanogroßen Barcodes, DNA-basierten Impfstofftechnologien sowie von strukturellen Proben mit multiplexierten molekularen Sensorprozessen spielen sollen. Größen, Formen sowie weitere für die ExperimentalphysikerInnen unsichtbare konformative Details wie die Größe der Hohlräume und der Grad der Rückfaltung der Äste wurden in Wien durch Computersimulationen analysiert. Um die komplexe Struktur von DNA-Einheiten zu beschreiben, verwendete die Gruppe ein Monomer-aufgelöstes Modell mit sorgfältig ausgewählten Wechselwirkungen, um die Gleichgewichtseigenschaften der DNA in physiologischer Lösung nachzuahmen. Die ausgezeichnete Übereinstimmung zwischen Experimenten und Simulationen für die Dendrimer-Eigenschaften bestätigt die verwendeten theoretischen Modelle und ebnet den Weg für die weitere Untersuchung der Eigenschaften von Nanokäfigen und ihrer Anwendungen als Nanoteilchen-Träger und als Bausteine für die Entwicklung biokompatibler künstlicher Materialien.
Die Forschung wird vom Österreichischen Fonds zur Förderung der wissenschaftlichen Forschung (FWF) unter der Projektnummer I 2866-N36 und von der Deutschen Forschungsgemeinschaft (DFG) unter der Projektnummer STI 664/3-1 gefördert.
Originalpublikation in "Nanoscale":
Clemens Jochum, Nataša Adžić, Emmanuel Stiakakis, Thomas L. Derrien, Dan Luo, Gerhard Kahl, and Christos N. Likos: Structure and stimuli-responsiveness of all-DNA dendrimers: theory and experiment, Nanoscale (2018).
DOI: 10.1039/C8NR05814H
Wissenschaftlicher Kontakt
Univ.-Prof. Dipl.-Ing. Dr. Christos N. Likos
Fakultät für Physik
Universität Wien
1090 - Wien, Sensengasse 8/15
+43-1-4277-732 30
christos.likos@univie.ac.at
> see press report
> see corresponding newspaper article in derStandard.at
> see corresponding article at science.orf.at